Biological nanopores for single-molecule analysis has been regarded as one of the most promising methods for inexpensive, high-speed and label-free DNA sequencing. Recently, a study from Prof. Yitao Long’s research group, East China University of Science and Technology (ECUST) demonstrated an advance in discriminating oligonucleotides by aerolysin nanopores. The paper entitled “Discrimination of oligonucleotides of different lengths with a wild-type aerolysin nanopore” has been published in Nature Nanotechnology. (http://www.nature.com/nnano/journal/vaop/ncurrent/full/nnano.2016.66.html)
The basic idea of nanopore sequencing is simple: while a single strand of DNA passes through a biological nanpore under the electric field, the different nucleotides (A, T, C, G) would cause four distinguishable blockade current signals. Therefore, the sequence information of a ssDNA can be read quickly without sample amplifying. However, reading of the bases from a DNA molecule in a nanopore has been hampered by the fast translocation speed of DNA together with the fact that several nucleotides contribute to the recorded signal. In the past two decades, in order to construct a suitable nanoscale pore for ssDNA detection, both pore-forming proteins and solid-state materials (SiNx, graphene, DNA origami, MoS et al) has been involved to fabricate a nanopore.
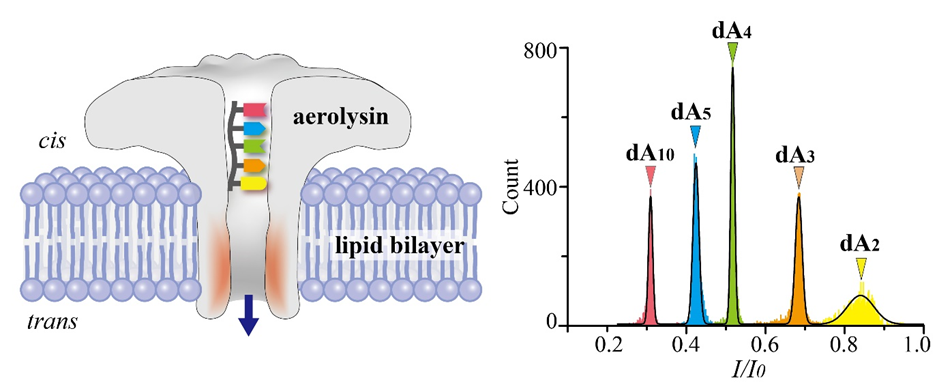
Researchers in ECUST's Laboratory of Analytical Biomicro System, which is leaded by Yitao Long reported a wild-type (WT) aerolysin as an excellent candidate for nucleic acid analysis for the first time. Chan Cao and her colleagues demonstrated that WT aerolysin nanopores exhibited an exceptionally high current separation for mixing dAn (dAn (n = 2, 3, 4, 5 and 10), the translocation speed is at approximately 0.3–2.0 ms nt–1, which is nearly three orders of magnitude slower than that through a WT α-haemolysin pore (3.3 μs nt−1 at 20 °C). Moreover, the results indicated that the wild-type aerolysin nanopores can distinguish individual oligonucleotides from mixtures and can monitor the stepwise cleavage of oligonucleotides by exonuclease I.
These exciting results made it possible that a direct detection of all individual nucleotides by using simple aerolysin-based approach if employed with high-bandwidth instruments. The excellent current separation enables aerolysin to achieve direct discrimination of single nucleotide variations, epigenetic modifications, DNA damage, single-molecule mass spectrometer and to examine the activity of nucleases.
This work was supported by the National Natural Science Foundation of China (grant nos 21327807, 21421004), the 111 Project (grant no. B16017) and National Basic Research Program of China (973 Program) (Grant no. 2013CB733700).




